The IDT Core offers many strategies for targeting your gene of interest, and since every gene is unique, each strategy is custom tailored to your gene.
These examples show the basic design for various targeting methods we use, and give a general overview of how your gene is modified.
3 Knockout (KO) Allele Strategies
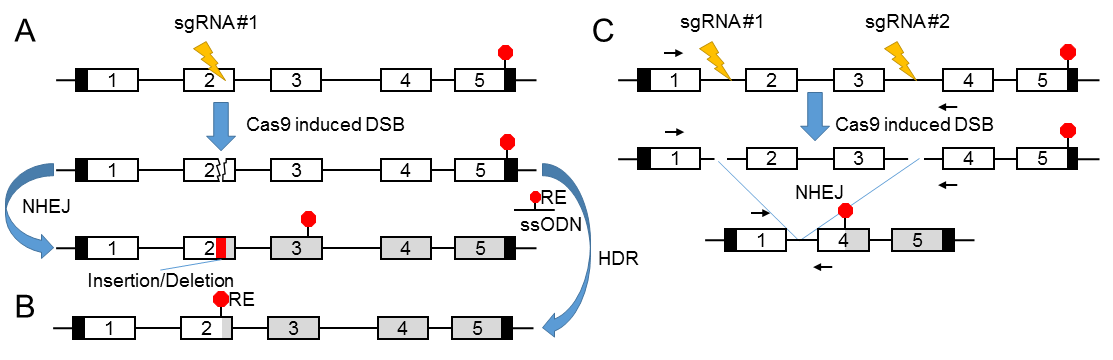
A) Strategy using a single sgRNA. Repair of the double strand break (DSB) by non-homologous end joining (NHEJ) may lead to small indels, which may create a frameshift and a premature stop codon.
B) Strategy using a single sgRNA and an ssODN template for repair using HDR.
C) Strategy using a two sgRNAs repair of the two DSBs and deletion of the intervening sequence leading to a large deletion and KO allele. Coding exons are represented by open boxes, untranslated region (UTR) by black boxes, coding sequence following a premature stop codon by grey boxes. Stop codons are represented by red octagons, and yellow lightning bolts represent the sites of DSBs induced by the Cas9-sgRNA complex. Genotyping primers flanking the large deletion in C are represented by small black arrows. RE: restriction site sequence.
Point Mutations
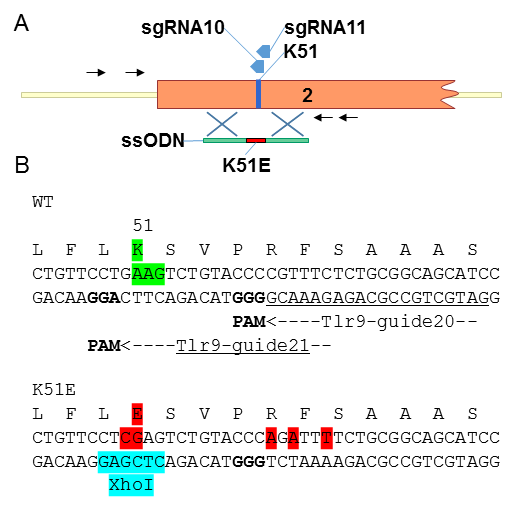
(A) Schematic of the genomic region encoding the lysine 51 of Tlr9. Exon 2 is in orange box, the position of K51 and of the sgRNA target sequence are depicted. Specific primers for PCR and nested primers for sequencing are represented by black arrows. Blue crosses represent the region of homology between the ssODN and the genomic sequence.
(B) Details of the substitutions in the ssODN that are introduced to create the K51E allele. The sgRNA target sequences are named and represented by arrows above and bellow the WT sequence, the PAM sequences are in bold, substitution in the ssODN are in red.
Conditional KO
For cKO, a 2 sgRNA with 2 ssODN strategy will be used to insert LoxP sites flanking critical exons.
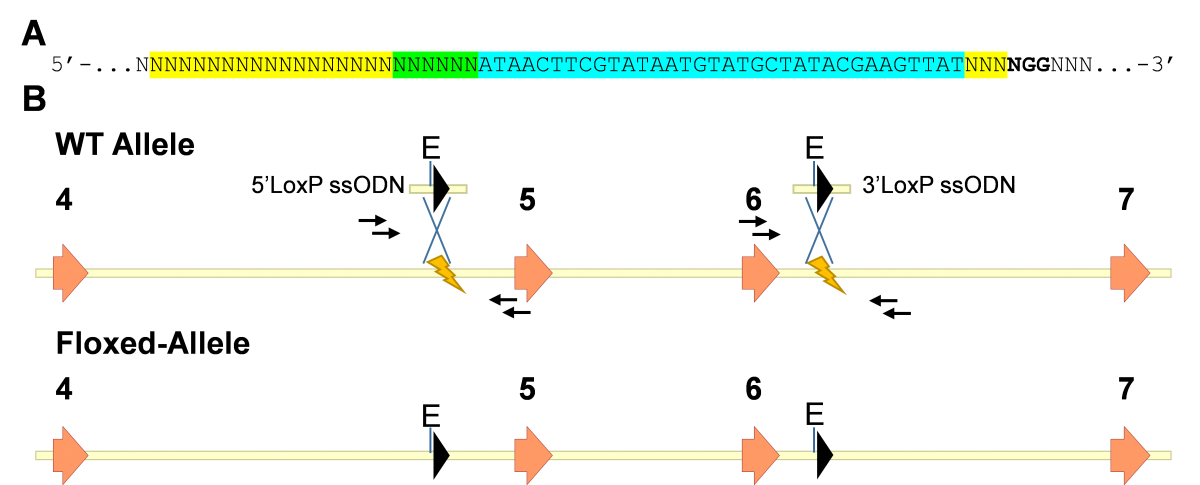
A) Structure of ssODN to introduce LoxP sites to generate conditional allele. All of the ssODN to introduce LoxP sites will have the same structure. The sequence for a LoxP site (34 bp, cyan) and a restriction enzyme (6 bp, green) will be introduced at the position of the DSB induced by Cas9, which is 3 bp upstream of the PAM sequence (NGG, bold), disrupting the sgRNA target sequence (yellow) and preventing further editing of the allele containing the LoxP by Cas9. Sequence of homology for HDR will be the 80 bp both upstream and downstream of the DSB position for a total of 200 bp (80 + 6 + 34+ 80).
B) Schematic for cKO, a generic gene is represented. Coding exons are orange arrows. The yellow lightning bolts represent the sites of DSBs induced by the Cas9-sgRNA complex. LoxP sites will be introduced by HDR at the sites of DSBs, using 2 ssODN (above WT allele). “X” between ssODNs and the WT allele represent recombination events that will lead to the generation of the floxed allele (bottom). LoxP sites are represented by black arrowheads. Genotyping primers flanking the LoxP sites are represented by small black arrows. “E” represents EcoR1 restriction sites.
